We are happy to announce the [Insight Toolkit (ITK)](https://itk.org) 5.4 Release Candidate 4 is available for testing! :tada: ITK is an open-source, cross-platform toolkit for N-dimensional scientific image processing, segmentation, and registration.
🔦 Highlights
-------------------------
The Insight Toolkit (ITK) has further enhanced its DICOM capabilities in the latest release, building on over 25 years of extensive clinical usage and application. DICOM (Digital Imaging and Communications in Medicine) is a valuable standard handling, storing, printing, and transmitting information in medical imaging. It includes a wide range of medical data types and allows for various imaging modalities and workflow information, posing significant challenges due to its extensive support for diverse medical content and variations in vendor implementations and adherence to the standard.
This release introduces expanded support for additional modality features and crucial spatial metadata for Secondary Capture images. ITK significantly improves the way medical imaging data is processed and interpreted and ensures robust support for diverse DICOM applications.
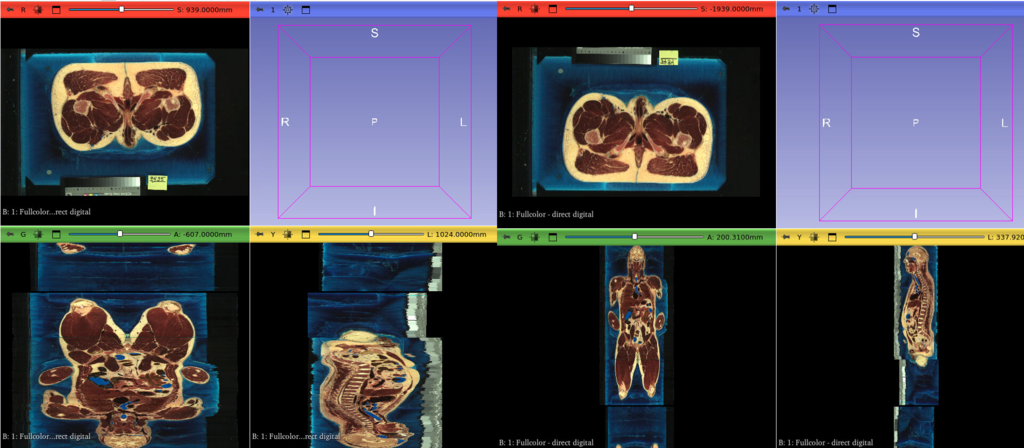
*The impact of spatial metadata handling on the NLM Visible Human cryomacrotome anatomic secondary capture images, [available in the NIH Imaging Data Commons](https://portal.imaging.datacommons.cancer.gov/explore/filters/?collection_id=nlm_visible_human_project), when visualized in [3D Slicer](https://slicer.org). Left: before ITK v5.4rc04, Right: after. Screenshots courtesy Steve Pieper.*
The development of ITK’s DICOM support is a testament to a robust community-driven effort involving ITK developers, maintainers from DICOM library projects such as GDCM and DCMTK, and curators of the DICOM standard. This release includes collaborative contributions from notable community members including Mikhail Isakov, Jon Haitz Legarreta Gorroño, Sean McBride, Martin Hoßbach, Mathieu Malaterre, Michael Onken, Steve Pieper, Andras Lasso, David Clunie, and Andrey Fedorov.
This release candidate also expands on our support for elegant, performant, modern C++. For example, specializations of `std::tuple_size ` and `std::tuple_element` for `itk::ImageRegion` in order to support C++17 structured bindings enable compile-time optimized statements to provide a multidimensional region's index and size:
cxx
auto [index, size] = image.GetRequestedRegion();
For more information on ITK 5.4's modern C++ support, see [the Release Candidate 1 release notes](https://docs.itk.org/en/latest/releases/5.4rc01.html).
Moreover, this release candidate extends the toolkit's sustainability and Python support through Stable ABI Python wheels. This is made possible by upgrades to SWIG and [scikit-build-core](https://scikit-build-core.readthedocs.io/en/latest/), the modern Python packaging standard evolution of scikit-build classic. Python 3.11 wheels will be recognized by `pip` and work with Python 3.11, 3.12, 3.13, 3.14, etc. While we also provide cross-platform wheels for Python 3.8-3.10, we can only use the Stable ABI with Python 3.11 because it is required for [`itk`'s NumPy support](https://docs.itk.org/en/latest/learn/python_quick_start.html#itk-and-numpy).
ITK Remote Modules now also have GitHub Action-driven mac ARM / Apple Silicon Python wheel generation support. While a Remote Module *setup.py* file is still supported in ITK 5.4, migration to a scikit-build-core *pyproject.toml* file is encouraged. One important advantage is the generation of Stable ABI wheels for Python 3.11+. To migrate to scikit-build-core, use [this pyproject.toml template](https://github.com/InsightSoftwareConsortium/ITKModuleTemplate/blob/main/%7B%7Bcookiecutter.project_name%7D%7D/pyproject.toml) and remove the *setup.py* file.
ITK 5.4 contains many additional improvements; highlights can be found below along with a more detailed changelog. For a summary of changes that continue our sustainability evolution with Web3 testing data, see the [5.4 Release Candidate 2 release notes](https://docs.itk.org/en/latest/releases/5.4rc02.html).
💾 Download
-------------
**Python Packages**
Install [ITK Python packages](https://docs.itk.org/en/latest/learn/python_quick_start.html) with:
bash
pip install --upgrade --pre itk
**Guide and Textbook**
- [InsightSoftwareGuide-Book1-5.4rc04.pdf](https://github.com/InsightSoftwareConsortium/ITK/releases/download/v5.4rc04/InsightSoftwareGuide-Book1-5.4rc04.pdf)
- [InsightSoftwareGuide-Book2-5.4rc04.pdf](https://github.com/InsightSoftwareConsortium/ITK/releases/download/v5.4rc04/InsightSoftwareGuide-Book2-5.4rc04.pdf)
**Library Sources**
- [InsightToolkit-5.4rc04.tar.gz](https://github.com/InsightSoftwareConsortium/ITK/releases/download/v5.4rc04/InsightToolkit-5.4rc04.tar.gz)
- [InsightToolkit-5.4rc04.zip](https://github.com/InsightSoftwareConsortium/ITK/releases/download/v5.4rc04/InsightToolkit-5.4rc04.zip)
**Testing Data**
Unpack optional testing data in the same directory where the Library Source is unpacked.
- [InsightData-5.4rc04.tar.gz](https://github.com/InsightSoftwareConsortium/ITK/releases/download/v5.4rc04/InsightData-5.4rc04.tar.gz)
- [InsightData-5.4rc04.zip](https://github.com/InsightSoftwareConsortium/ITK/releases/download/v5.4rc04/InsightData-5.4rc04.zip)
**Checksums**
- [MD5SUMS](https://github.com/InsightSoftwareConsortium/ITK/releases/download/v5.4rc04/MD5SUMS)
- [SHA512SUMS](https://github.com/InsightSoftwareConsortium/ITK/releases/download/v5.4rc04/SHA512SUMS)
✨ Features
------------
Python
- Wrapping for `itk.PhasedArray3DSpecialCoordinatesImage`
- Better support for multi-component images in `image_from_vtk_image`
- `itk.imread` supports a `series_uid` kwarg for DICOM series selection
- TBB version updated to latest stable version, disabled on Intel macOS
- Python binaries for 3.8-3.11 across platforms
- Python 3.11 uses the Stable ABI -- works with Python 3.11+
- Python 3.7 is no longer supported
- Apple Silicon Remote Module GitHub Action wheels
- Updated to the latest version of scikit-build-core
- Import time improvements with `torch`
C++
- C++17 is now required
- Many style improvements for modern C++ and consistency
- GCC 13 support
- Name mangling prefix for third party libraries is configurable
- Update mangled 3rd-parties to use `MANGLE_PREFIX` CMake variable
- Many improvements to code coverage
- Enhanced NRRD and Nifti metadata support
- CMake `OPTIONAL_COMPONENTS` support
- Apply cmake-format for a consistent CMake style
- `get()` member function to `itk::SmartPointer`
- `itk::Size::CalculateProductOfElements()`, to compute number of pixels
- `Deref(T *)`, to ease dereferencing a pointer safely
- `itk::ShapedImageNeighborhoodRange` support C-array of offsets (by C++17)
- Add `itk::Copy(const T & original)`, which simply returns a copy
- Make `itk::ImageRegion` trivially copyable, remove inheritance (FUTURE)
Performance
- Use index/point transforms without bounds checking
- Improved SSE2 detection
- Many improvements to how locks are handled
- Major `itk::SpatialObject` performance improvements
Documentation
- New GitHub Action to check spelling
- Doxygen formatting cleanup
- Doxygen spelling fixes
- Doxygen Insight Journal links are consistent
- Many Doxygen improvements to the content
- Software Guide updated for style modernization
- Change the Insight Journal handle links to insight-journal links
- Replace `itkTypeMacro` with `itkOverrideGetNameOfClassMacro`
Remote module updates
New modules:
- [FastBilateral](https://hackmd.io/I4AtOFiKSI6rvHKm1fYmeA) - A Fast Approximation to the Bilateral Filter for ITK. [Insight Journal article](https://insight-journal.org/browse/publication/692).
Updated modules:
- [BSplineGradient](https://github.com/InsightSoftwareConsortium/ITKBSplineGradient.git)
- [BoneMorphometry](https://github.com/InsightSoftwareConsortium/ITKBoneMorphometry.git)
- [Cleaver](https://github.com/SCIInstitute/ITKCleaver.git)
- [Cuberille](https://github.com/InsightSoftwareConsortium/ITKCuberille.git),
- [CudaCommon](https://github.com/RTKConsortium/ITKCudaCommon.git)
- [FPFH](https://github.com/InsightSoftwareConsortium/ITKFPFH.git)
- [GenericLabelInterpolator](https://github.com/InsightSoftwareConsortium/ITKGenericLabelInterpolator.git)
- [HASI](https://github.com/KitwareMedical/HASI.git)
- [HigherOrderAccurateGradient](https://github.com/InsightSoftwareConsortium/ITKHigherOrderAccurateGradient.git)
- [IOMeshSTL](https://github.com/InsightSoftwareConsortium/ITKIOMeshSTL.git)
- [IOMeshSWC](https://github.com/InsightSoftwareConsortium/ITKIOMeshSWC.git)
- [IOScanco](https://github.com/KitwareMedical/ITKIOScanco.git)
- [LabelErodeDilate](https://github.com/InsightSoftwareConsortium/ITKLabelErodeDilate.git)
- [MeshToPolyData](https://github.com/InsightSoftwareConsortium/ITKMeshToPolyData.git)
- [MinimalPathExtraction](https://github.com/InsightSoftwareConsortium/ITKMinimalPathExtraction.git)
- [Montage](https://github.com/InsightSoftwareConsortium/ITKMontage.git)
- [MorphologicalContourInterpolation](https://github.com/KitwareMedical/ITKMorphologicalContourInterpolation.git)
- [ParabolicMorphology](https://github.com/InsightSoftwareConsortium/ITKParabolicMorphology.git)
- [RANSAC](https://github.com/InsightSoftwareConsortium/ITKRANSAC.git)
- [RLEImage](https://github.com/KitwareMedical/ITKRLEImage.git)
- [RTK](https://github.com/RTKConsortium/RTK.git)
- [Shape](https://github.com/SlicerSALT/ITKShape.git)
- [SimpleITKFilters](https://github.com/InsightSoftwareConsortium/ITKSimpleITKFilters.git)
- [SplitComponents](https://github.com/InsightSoftwareConsortium/ITKSplitComponents.git)
- [Strain](https://github.com/KitwareMedical/ITKStrain.git)
- [TextureFeatures](https://github.com/InsightSoftwareConsortium/ITKTextureFeatures.git)
- [Thickness3D](https://github.com/InsightSoftwareConsortium/ITKThickness3D.git)
- [TubeTK](https://github.com/InsightSoftwareConsortium/ITKTubeTK.git)
- [Ultrasound](https://github.com/KitwareMedical/ITKUltrasound.git)
- [VkFFTBackend](https://github.com/InsightSoftwareConsortium/ITKVkFFTBackend.git)
- [WebAssemblyInterface](https://github.com/InsightSoftwareConsortium/itk-wasm.git)
Third party library updates
- dcmtk
- eigen
- gdcm
- googletest
- kwsys
- minc
- meta-io
- nifti
- pygccxml
- vxl
- zlib-ng
🙏 Congratulations
------------------------
Congratulations and **thank you** to everyone who contributed to this release.
Of the *59 authors* who contributed since v5.3.0, we would like to specially recognize the new contributors:
*Nicklas Larsson, huangjxbq, Sankhesh Jhaveri, adrinkwater, FabioLolix, Vaibhaw, Ningfei Li, Max Aehle, Noah Egnatis, Federico Zivolo, Patrick Linnane, LAURENDEAU Matthieu, Shreeraj Jadhav, Shengpeng YU, Fernando Bordignon, Andras Lasso, Bernhard Froehler, Thomas BAUDIER, Matthieu LAURENDEAU, Fabian Wenzel, Mikhail Polkovnikov, Pritam Rungta, Florian de Gaulejac, Ramon Emiliani, Martin Hoßbach, Sadhana Ravikumar, and Gabriel Chartrand.*
🗣️ What's Next
---------------
This is the final release candidate before v5.4.0. An issue following the tagging of v5.4rc03 predicated the publication of this release. Please try out the current release candidate, and discuss your experiences at [discourse.itk.org](https://discourse.itk.org). Contribute with pull requests, code reviews, and issue discussions in our [GitHub Organization](https://github.com/InsightSoftwareConsortium).
**Enjoy ITK!**