- from pr 130
- commits from: banesullivan
- review from: lheagy
Add vtki support to make using VTK data objects more Pythonic
banesullivan recently added a ton of new features to [`vtki`](https://github.com/akaszynski/vtki) (the VTK interface Python package) that help make using just about any VTK data objects more intuitive/Pythonic. If `vtki` is available in your Python environment then these changes make using VTK data objects way easier. Here's a screenshot of what this currently looks like in a Jupyter notebook (creates static renderings but can also be interactive in a separate pop-up window).
Also check out [this notebook](https://github.com/OpenGeoVis/PVGeo-Examples/blob/master/3.0%20-%20Using%20the%20vtkInterface%20Package.ipynb) to see more ways to use [`PVGeo`](https://github.com/OpenGeoVis/PVGeo), `discretize`, and [`vtki`](https://github.com/akaszynski/vtki). Below is a screenshot of a simple use case:
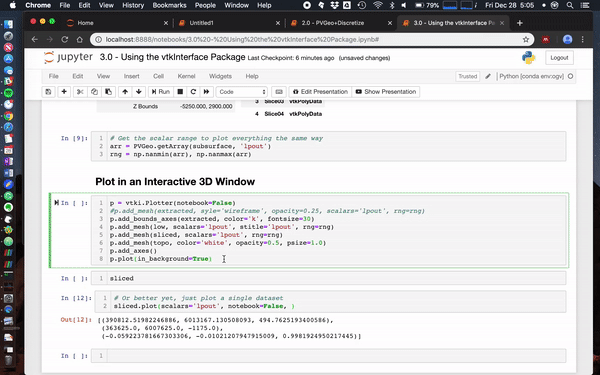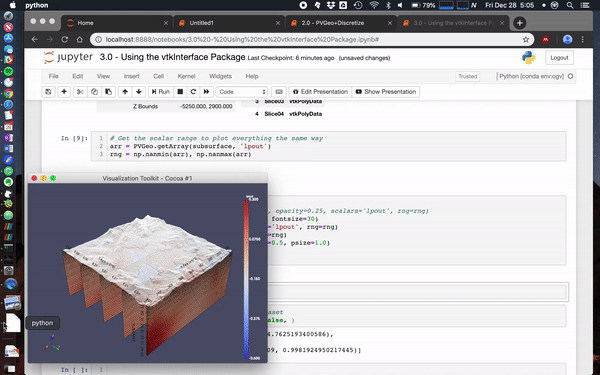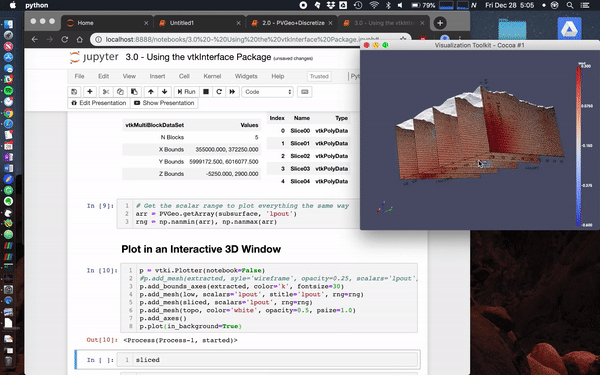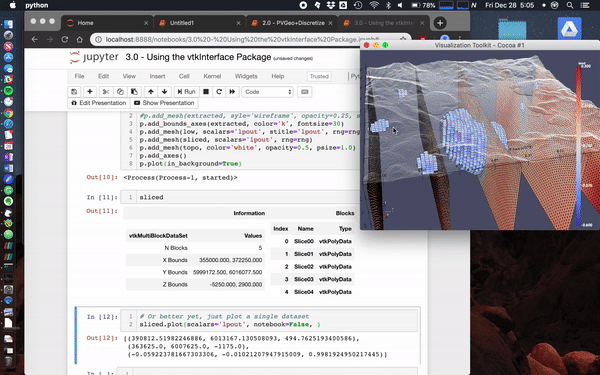
<img width="757" alt="screen shot 2018-12-28 at 2 27 21 pm" src="https://user-images.githubusercontent.com/22067021/50525947-ba6eff80-0aac-11e9-8b1d-dfbc770fe3c9.png">
Simple example
py
import discretize
import numpy as np
Create a simple TensorMesh
h1 = np.linspace(.1, .5, 3)
h2 = np.linspace(.1, .5, 5)
h3 = np.linspace(.1, .5, 3)
mesh = discretize.TensorMesh([h1, h2, h3])
Convert to VTK object and use vtki to render it
mesh.toVTK().plot(notebook=False)
Fancy example
Here we load the [**Laguna del Maule Bouguer Gravity**](http://docs.simpeg.xyz/content/examples/04-grav/plot_laguna_del_maule_inversion.html#sphx-glr-content-examples-04-grav-plot-laguna-del-maule-inversion-py) example from the SimPEG docs.
This data scene was produced from the [Laguna del Maule Bouguer Gravity](http://docs.simpeg.xyz/content/examples/04-grav/plot_laguna_del_maule_inversion.html#sphx-glr-content-examples-04-grav-plot-laguna-del-maule-inversion-py) example provided by [Craig Miller](https://github.com/craigmillernz) (Maule volcanic field, Chile. Refer to Miller et al 2016 EPSL for full details.)
> Miller, C. A., Williams-Jones, G., Fournier, D., & Witter, J. (2017). 3D gravity inversion and thermodynamic modelling reveal properties of shallow silicic magma reservoir beneath Laguna del Maule, Chile. Earth and Planetary Science Letters, 459, 14–27. https://doi.org/10.1016/j.epsl.2016.11.007
py
import shelve
import tarfile
import discretize
f = discretize.utils.download(
"https://storage.googleapis.com/simpeg/laguna_del_maule_slicer.tar.gz"
)
tar = tarfile.open(f, "r")
tar.extractall()
tar.close()
with shelve.open('./laguna_del_maule_slicer/laguna_del_maule-result') as db:
mesh = db['mesh']
Lpout = db['Lpout']
mesh = discretize.TensorMesh.copy(mesh)
models = {'Lpout':Lpout}
mesh.toVTK(models).plot()
Usage Notes
Since `vtk` and `vtki` are not required dependencies you'll need to make sure you install them to your Python environment. Pip install for `vtki` should do the trick but Windows folks might need to install `vtk` from conda seperately. Also this is Python 3 friendly!
sh
pip installl vtki