Release notes
Base pair lines
It is common in DNA nanotech figures to draw lines orthogonal to the DNA strand lines representing base pairs. Although this would not help when designing a scadnano design, it would help when making figures. By enabling View --> Base pairs --> Base pair lines:

Lines between base pairs are drawn. In combination with turning off the view of helices (View --> Helices --> Show main view helices), this makes it easy to make "figure quality" diagrams in scadnano:

Domain/Extension/Loopout colors
This is another feature useful for making paper-figure-quality diagrams with scadnano.
Individual `Domain`'s, `Extension`'s, and `Loopout`'s now have a `color` field. If specified, then this overrides the field `Strand.color` in the display. In the following image, the strand's color is blue, which is the color of the bottommost domain (which has no color specified) and is also the color of all the crossovers. Every other domain/extension/loopout has its own custom color specified:

They can be specified in the Python package, and manually by right-clicking:


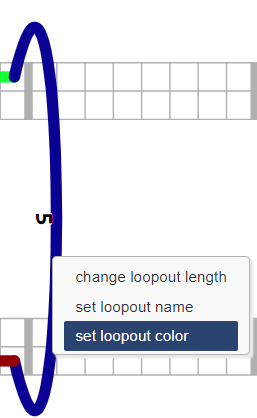
Note that although domain/extension/loopout color is optional, strand color is required.
Select all in helix group
Strands from different helix groups cannot be copied or moved. To make it easier to select all strands in a single group (rather than all strands in the whole design), there is now command "Edit --> Select All in Helix Group" (keyboard Ctrl+Shift+A) that selects all strands in the current helix group.
Commits
- 220d670: bumped version (David Doty) [833](https://github.com/UC-Davis-molecular-computing/scadnano/pull/833)
- 07f1cc8: closes 827: add "select all in current helix group" (Edwin Chang) [828](https://github.com/UC-Davis-molecular-computing/scadnano/pull/828)
- 0c5b9b6: Remove extra action and add keyboard shortcut (Edwin Chang) [828](https://github.com/UC-Davis-molecular-computing/scadnano/pull/828)
- e7b0e8b: Add tests for select all strands in helix group (Edwin Chang) [828](https://github.com/UC-Davis-molecular-computing/scadnano/pull/828)
- 3d9c697: Fixed selection issues for strands in multiple groups (Ho-Chih) [828](https://github.com/UC-Davis-molecular-computing/scadnano/pull/828)
- 6c41702: closes 829: Optional base pair lines (David Doty) [831](https://github.com/UC-Davis-molecular-computing/scadnano/pull/831)
- ffcb86e: added support for displaying domains/loopouts/extensions with optional color field; still need to support setting colors in the web interface (David Doty) [832](https://github.com/UC-Davis-molecular-computing/scadnano/pull/832)
- 4d1dde0: closes 796: Domain colors (David Doty) [832](https://github.com/UC-Davis-molecular-computing/scadnano/pull/832)